Achieving Data Liquidity in the Cancer Community: Proposal for Coalition of All Stakeholders
Statement of Purpose
Rapid, seamless data exchange in cancer—throughout the continuum from research to clinical care—remains an unmet need with no clear path forward to a solution. We assert that there is a national urgency to find solutions to support and sustain the cancer informatics ecosystem (IOM, 2012c), and propose addressing this challenge through a coalition of diverse, interested stakeholders working in a precompetitive collaboration to achieve data liquidity. Such a coalition could work toward the goal of personalized cancer medicine, addressing the challenges in manageable, incremental steps.
The Opportunity
Personalized Cancer Medicine
Countless voices across the research and care enterprise have heralded the current era as having transformative potential for cancer treatment and prevention. Such transformation will be due in part to the unprecedented opportunities provided by molecular understanding of disease, leading to personalized (also known as gene-based, stratified, targeted, or precision) cancer medicine (AACR, 2011; NRC, 2011).
At the same time, thought leaders in biomedicine have sought to improve cancer care through the development of a “rapid learning health care system.” This system will link research and care into a seamless process by using “advances in information technology [IT] to continually and automatically collect and compile from clinical practice, disease registries, clinical trials, and other sources of information, the evidence needed to deliver the best, most up-to-date care that is personalized for each patient” (IOM, 2010b). Stakeholders have invested significant thought in conceptualizing the digital infrastructure necessary to support a rapid learning health care system (IOM, 2011).
At the intersection of personalized cancer medicine and the rapid learning health care system is the IT challenge: “Not all personalized medicine is molecular based, but the need to move to evidence-based personalized medicine requires the design of new information systems that enable the repurposing of data for multiple stakeholders” (Fenstermacher et al., 2011).
Full realization of personalized cancer medicine presumes the existence of information systems that allow for data liquidity. Data liquidity is defined herein as the rapid, seamless, secure exchange of useful, standards-based information among authorized individual and institutional senders and recipients. Ideally, such data liquidity would span a continuous cycle that supports basic discovery research, translational research, product development, clinical trials, comparative effectiveness research, health services research, and delivery of personalized cancer care and prevention interventions to patients. All of these domains are inextricably linked and face the same fundamental information-exchange challenge, albeit for different specific objectives.
What If…
The following personalized cancer medicine scenarios of the future illustrate the potential role that data liquidity could have in advancing research and care:
- All types of researchers in various disciplines—including basic science, drug discovery, clinical and translational, epidemiologic, and patient behavior—will have access to the requisite data to advance their research. They will have access to data needed to support discovery of molecular signatures that identify populations at risk, are predictive of response to therapy, provide prognosis and risk of recurrence information, and facilitate identification of new targets by drug developers that can be used for non-responders. They will have the real-time capability to communicate with the physicians providing clinical data and their patients participating in clinical trials. They will be able to quickly screen patients who may be clinically eligible for a particular clinical trial, assess the prevalence of specific mutations or combinations of mutations, and then identify the associated patients and their physicians for follow-up contact. This will be supported by access to analytics that identify and profile individuals with information available using data structures securely connecting multiple forms of molecular and clinical data.
- Clinicians will have immediate access to cross-sectional, trending, and performance data that may facilitate improved clinical decision making. Clinicians will be able to more rapidly identify patients at risk for disease and poor outcomes. By using validated cancer biomarkers to identify patients at risk, prevention and other early intervention steps will be taken, and interventions, orders, and results will be tracked in patient populations to provide a “closed loop” learning laboratory for improving approaches to promote wellness and prevent disease.
- Patients will have real-time access to information for finding reliable sources pertinent to their particular condition and finding expertise tailored to their individual needs. Patients will be able to connect their clinical information from their health provider to personal health record portals, enabling data integration and facilitating the use of non-traditional, consumer-controlled data sources. Patients will be able to view and choose to submit their data to their researchers/clinicians, to further link care to research.
- Product innovators will have the ability to query large amounts of phenotypic and molecular data, and rapidly scan patient populations for recruitment into clinical trials. They will be able to share molecular, clinical, laboratory, and imaging data seamlessly within multi-institution research collaborations. They will be able to facilely bridge clinical treatment data with clinical trials data, and track product performance pre- and postmarketing.
Data liquidity provides the capability to aggregate disparate sources of data (thereby creating “big data”) to be used in a meaningful way. The potential value of using big data includes a variety of productivity gains in health care. According to a recent McKinsey report, five big data levers could improve R&D productivity and create more than $100 billion in value: predictive modeling, statistical tools and algorithms to improve clinical trial design, analysis of clinical trials data, personalized medicine, and analysis of disease patterns (The McKinsey Global Institute, 2011). All five levers, to be sure, will require data liquidity.
The Challenges
At this critical juncture, biomedicine has no systematic means for rapid and efficient exchange of data so that it can be leveraged and converted into knowledge by anyone other than the original author. This digital dilemma is particularly apparent in cancer research, which has been a pioneer in advancing molecularly targeted approaches to treatment. However, these research advances are dependent on informatics to interpret complex, high-dimensional data. A lack of infrastructure and methodologies to connect and interpret this information may be slowing research progress.
Collecting the Right Data, and Collecting the Data “Right”
As individuals and institutions in biomedicine seek to implement the rapid learning health care system, a common concern is that the requisite data may not get collected in our current system, either in paper or electronic form, making the exchange from data “source” to data “output” highly challenging. Massive quantities of data cannot necessarily compensate for missing categories of data, and sheer quantity will not necessarily facilitate translation of data into knowledge. Rather, it is often what types of data and how they are collected that is of paramount importance. Data will only be as useful as the data elements used to comprise them. In the clinical setting, we assert that critical variables should be captured. For example, information about why a patient discontinued a drug may be of key importance to understanding the side-effect profile of that therapy, and may provide more information about the progression of disease in certain patient subpopulations. Yet, such information may not currently be gathered at all, or may be gathered in free-text form (rather than a structured format) that precludes effective analysis. Information about the ultimate clinical outcome for a patient may also be lost, as many electronic health records (EHRs) capture only what care was delivered, rather than patient response. In addition, as cancer patients move from active cancer treatment to primary and survivorship care, important information may not be captured or conveyed in this transition (IOM and NRC, 2006). Similarly, in the research setting, coding of genetic mutations and the terminology around complex molecular pathways is still in development and evolving, making it difficult to compare across studies.
As noted by a cancer center currently implementing a personalized medicine initiative: “The principles of data accuracy, completeness, time-relatedness, consistency, accessibility, and integrity are central themes of [Total Cancer Care] data management” (Fenstermacher et al., 2011). In that center, it has been essential to carefully delineate the protocols for the “right” data extraction, data transformation, semantic integration, data profiling, and data cleansing, especially because 11 institutions are participating in their Research Information Exchange.
Finally, it is important to recognize that the growing adoption of EHRs—even with associated tools for ubiquitous collection of patient-reported data—will not on its own incorporate the necessary research data elements into clinical care systems, nor will it address the fundamental problem of connectivity and data liquidity faced by the spectrum of cancer research that underpins many of the future scenarios delineated above.
Disincentives to Data Sharing
Widespread sharing of data in biomedicine has not been the norm because of a number of disincentives that are difficult to address individually. For example, privacy advocates have traditionally sought to maximize individual privacy and patient authorization for the use of clinical information. In addition, current federal privacy regulations promulgated under the Health Insurance Portability and Accountability Act impede health research and the exchange of patient data (IOM, 2009). Research incentives have focused on individual investigator achievement and publication rather than on collaboration enabled by data sharing, and publication is still the key to tenure and promotion for academic scientists (IOM, 2010a,c). In addition, health care providers have sought to gain competitive advantage from the knowledge gained from care of their own patient population, reflecting today’s economic pressures. Pharmaceutical companies have been protective of the intellectual property at the heart of their massive investment in new products, including reluctance to share data even on the failures in their pipeline because there has been little perceived incentive to do so (IOM, 2012b). More recently, patient advocacy organizations have sought to protect the financial potential of the member-data “asset” they possess. Finally, concerns over data security for the most personal information we possess have had a chilling effect on the kinds of large-scale data transfer that are taken for granted in industries such as banking and retail.
Even when data sharing is agreed to in principle, challenges remain in governance because issues about who should have access to the data and what protocols need to be followed may require extensive discussion and complex approaches to resolution, in ways that are not typical of other sectors.
Technical Obstacles
In spite of the dramatic advancements in information technology in recent years, technical obstacles still abound.
First is the sheer quantity of data to be managed. The biomedical enterprise is replete with big data of all kinds, such as biological, molecular, clinical, laboratory, pharmacy, and administrative data. Moreover, the overall pool of biomedical data is growing exponentially as the cost of genomic sequencing drops and the application of imaging technologies expands.
Second is the non-uniformity of the data. Much of the potentially shareable data are of varying quality, are non-conformant to standard vocabularies, and are often prohibitively laborious or impossible to translate from one discipline to another. As noted in the Institute of Medicine (IOM) workshop on precompetitive collaboration, “addressing inconsistent or otherwise non-comparable data requires standards and infrastructure that can be costly to develop” (IOM, 2010a). Ironically, the field is not challenged by an absence of standards; rather, it is challenged by the fact that multiple standards are available within different domains, none of which are universally adopted across disciplines.
Third is the absence of incentives for building interconnected systems. Software vendors and service providers have no incentive to promote interoperable systems that might undermine the proprietary nature of their commercial arrangements or limit investment in their services. IT solutions developed in academia, which might be intended for the public good, have tended to be homegrown, site-specific, and not scalable for the larger research enterprise.
Fourth is the “growing complexity of basic and clinical research in oncology, much of which hinges on deciphering the intricate networks of molecular pathways involved in the formation and progression of various cancers, as well as predicting patients’ likely responses to treatments aimed at the targets within those networks. For many assays performed to determine perturbations in molecular pathways, there are no accepted measurement techniques or scales to express findings. The increasing need to integrate genetics, genomics, and proteomics into new drug development requires data repositories and much more sophisticated information technology to analyze data” (IOM, 2010a). Managing and accessing data represents the tip of the informatics challenge: there is also a need for novel computational tools and algorithms to extract insight from these large, multidimensional datasets in order to convert data to information, and make the information actionable. The transformation of data to knowledge requires not only tools, but an expanded collection of individuals trained to build that bridge between data and knowledge in biomedicine (IOM, 2012a).
Health IT
All the challenges above exist at a time when a national effort is underway in the United States to move toward an electronic health care environment. In 2011, the Centers for Medicare & Medicaid Services (CMS) began reimbursing health care providers and hospitals for the adoption of certified systems that support meaningful use of electronic health records (Blumenthal, 2010). The criteria needed to qualify for reimbursement center on reporting requirements and quality measures, but they do not include provisions for such systems to be able to exchange and aggregate data across an interconnected system. The absence of such provisions is a disincentive for the health informatics and IT-developer communities to address the challenges of robust information exchange. Such concerns have been noted by the President’s Council of Advisors on Science and Technology, who called for the development of a universal exchange language to enable information exchange across institutional boundaries, as well as for CMS to shift the focus of meaningful use guidelines to support a more comprehensive ability for health information exchange (PCAST, 2010). In short, while the foundation is being laid for the transition of the health care community to an electronic ecosystem, critical work is still needed to enable an environment that incentivizes the use of EHRs for research, integrates technologies from research to care, and connects information across institutions in a learning environment. The Certification Commission on Health Information Technology has set criteria for EHRs in the area of clinical research and oncology that provide a minimum standard set of elements, but these have not been adopted by the majority of EHR systems. Without these critical clinical data points represented as discrete structured elements, this work will be far less successful.
These concerns are reinforced by a recent report from the Bipartisan Policy Center, which noted that although there is considerable momentum toward adoption of EHRs, the “level of health information exchange in the U.S. is extremely low” (Bipartisan Policy Center Task Force on Delivery System Reform and Health IT, 2012).
The Role of Government
To date, government entities such as the National Institutes of Health (NIH), the Food and Drug Administration (FDA), and CMS have not routinely required the adoption of IT standards necessary to address the digital dilemma in biomedicine, nor have they developed a consistent framework for data exchange throughout the health care enterprise.
The National Cancer Institute’s (NCI’s) caBIG® initiative developed a first-generation national framework for data exchange. The NCI’s Board of Scientific Advisors Ad Hoc Working Group noted in its 2011 report: “Perhaps the greatest impact of the caBIG® program on cancer research has been to gather several communities around a virtual table to help create and manage community-driven standards for data exchange and application interoperability. The development of a semantic infrastructure that allows data to be harmonized across cancer centers is widely perceived to be one of the most important contributions of the caBIG® program” (NCI,
2011). Components of the caBIG® program underpin the data exchange used in the successful models of collaboration described below.
Exemplars of Data Exchange in Research
Success in data exchange is not a one-size-fits-all proposition, and must therefore be tailored to specific project and institutional needs. A few models described below demonstrate how common digital frameworks for data exchange can function reasonably well across diverse efforts.
- The Cancer Genome Atlas (TCGA) is a several-hundred-million-dollar collaborative project of the NCI and National Human Genome Research Institute, encompassing government and academia, seeking to “facilitate future discovery of pharmaceutical and diagnostic targets in cancer by generating genome characterization data on 20 tumor types” (IOM, 2010a). The massive amounts of multidimensional genomic data that TCGA is generating are coded to standards-based, common data elements and shared through open-source infrastructure.
- The Biomedical Research Integrated Domain Group (BRIDG) is a collaborative effort involving the Clinical Data Interchange Standards Consortium, the Health Level 7 Regulated Clinical Research Information Management Technical Committee, the NCI, and the FDA (Biomedical Research Integrated Domain Group, 2012), to provide a standard information model interconnecting clinical care data with clinical trial data. The BRIDG model is now being used by several biotechnology and pharmaceutical companies, allowing them to leverage clinical information for regulatory submissions. The common representation of information also permits the comparison of information among independent trials. In addition to the value to product innovators in expediting such submissions, the BRIDG capabilities exemplify the rapid learning health care system by allowing clinical research to inform clinical care and vice versa.
- The I-SPY 2 TRIAL (Investigation of Serial studies to Predict Your Therapeutic Response with Imaging And moLecular analysis 2) is pioneering an adaptive clinical trial design intended to be more flexible than traditional trials, proceed more rapidly, and experiment with new models of collaboration. The I-SPY 2 TRIAL is sponsored by the Foundation for the NIH’s Biomarkers Consortium, a partnership involving the FDA, the NIH, leading academic medical centers, pharmaceutical companies, and nonprofit and patient advocacy groups (I-SPY 2, 2012). The I-SPY 2 TRIAL uses metadata based on international standards (including BRIDG), as well as open-source infrastructure (including caTissue, caArray, and caIntegrator). Using this interoperable framework, the trial has been able to connect and share multidimensional data (clinical, molecular, and in vivo imaging), bridge information between the research and care settings, and share clinical research data with participants.
While these examples illustrate the feasibility of adopting standards and a common framework to facilitate exchange of complex multidimensional data, they are all one-off settings, confined to their own specific missions. It remains a national challenge to leverage these successes and gain synergy by building on, coordinating, and disseminating such data-exchange capabilities.
Proposed Pathway to a Solution
The IOM has conducted workshops focused on precompetitive collaborations in cancer research; on the rapid learning health care system in cancer; and on digital infrastructure necessary for a rapid learning health care system, which cover the multiple dimensions needed to achieve data liquidity. We therefore propose a solution at the intersection of these three subjects that integrates the insights from previous IOM workshops to address the informatics needs of the cancer community.
A Coalition of All Stakeholders
Because the problem of data liquidity cuts across the entire research spectrum, it will most likely require a solution that encompasses all stakeholders. As noted above, previously disparate sectors of the research and clinical communities have a common need for data exchange, and are seeking knowledge that affects the overall health care enterprise.
We envision a nonprofit membership organization composed of a broad range of commercial, academic, and consumer stakeholders in the life sciences and health care. The stakeholders could include the broadest possible representation: basic research scientists; clinical researchers; bioinformatics and IT experts; subject-matter experts from a wide range of disciplines; clinicians, biostatisticians, comparative-effectiveness research experts, health services researchers, epidemiologists, patients and advocacy organizations, payers, and indeed all types of developers and “consumers” of the digital framework.
The vision of such a coalition could reflect the virtuous cycle envisioned by the rapid learning health care system—information-based biomedicine in which all data about clinical care are applied to fuel and facilitate research and development of new treatments, and all research data are rapidly translated into knowledge to improve clinical care.
Coalition Principles
Members of the coalition could come together collaboratively to agree on core principles that might include some or all of the following:
- Research and clinical care can benefit from the collection and analysis of all relevant (and potentially relevant) information, recorded in standards-based formats and curated through the appropriate governance structures.
- Biomedical innovation and achievement of personalized cancer care can be accelerated and improved by facilitating connectivity and seamless data exchange among research and care collaborators.
- Open information technology frameworks, which define standards by which technology components can interoperate, provide such capability through open interfaces, enabling researchers to seamlessly capture, aggregate, integrate, analyze, interpret, and transmit data.
- A precompetitive setting in a connected biomedical community that benefits all health care stakeholders is the best setting for progress, where all participants are free to contribute and partake of components of the framework that would otherwise be inaccessible.
- Open-technology frameworks, which can facilitate the interface between open-source and/or commercial components, must be shared freely.
Coalition Operational Strategy
The operational strategy for the coalition could act on the insights gleaned from the 2011 IOM workshop for addressing digital infrastructure for the rapid learning health care system (IOM, 2011). These insights address the complex characteristics of the requisite digital infrastructure, including conflicting and diverse requirements, continuous evolution, heterogeneity with changing elements, and constant iteration.
The coalition could start with concretely defined projects supported by a user with a specified need. The solutions could be built in small increments and shared with the users at early stages of development. The users, in turn, could provide rapid feedback that could be used to iterate the next cycle of development. In that way, the solution would likely meet the users’ specified needs. Each of the solutions could serve as a building block that could be reused for future users, extended as needed, and aggregated to achieve more complex capabilities.
The viability of this strategy is evidenced by its demonstrable success in creating large, complex systems, and its use by the Internet Engineering Task Force in the ongoing development of the Internet. The strategy gives priority to working systems that can be quickly implemented, embracing a belief in what is called “rough consensus and working code” (Internet Engineering Task Force, 2012).
It is also assumed that small successes will garner increased attention from the clinical and research communities, drawing in more and more participants.
Coalition Governance
Nonprofit status for the coalition may be the most appropriate model to enable and ensure the honest broker role. Within the nonprofit context, either 501(c)(3) or 501(c)(6) tax-code status could be envisioned. The latter designation, comparable to a “business league” or Chamber of Commerce, may be preferable because it permits greater flexibility in operations, including the option of accepting financial resources beyond charitable donations. In addition, the 501(c)(6) status also has the potential advantage of emphasizing the desire to avoid competition with the private sector because by definition such an entity is not permitted to compete with commercial
firms that could perform coalition activities. (See http://www.irs.gov/charities/nonprofits/ (accessed January 4, 2012): “To be exempt, a business league’s activities must be devoted to improving business conditions of one or more lines of business as distinguished from performing particular services for individual persons. No part of a business league’s net earnings may inure to the benefit of any private shareholder or individual and it may not be organized for profit to engage in an activity ordinarily carried on for profit (even if the business is operated on a cooperative basis or produces only enough income to be self-sustaining).”)
Another model for consideration is the “B Corporation,” a legal framework for companies that seek to benefit society as a whole rather than having to concentrate on maximizing profits (B Corporation, 2012). Seven states have recently passed legislation to permit such “benefit” corporations as an alternative business model (Loten, 2012). The potential advantage of this approach is the ability to attract social entrepreneurs, and to emphasize the goals of the entity as a social priority to improve health care while recognizing that financial sustainability is still a requisite.
Coalition Objectives
The coalition could seek to achieve the following:
- Catalyze and help nurture a community to develop and make available, precompetitively, an open digital framework for biomedicine.
- Ensure that the open digital framework stays current with all technological advances.
- Ensure that all biomedical organizations have access to the open digital framework so they can achieve their goals for improved patient care and more productive research.
- Help to support a flourishing ecosystem of biomedical organizations that can fuel each other’s activities through frictionless flow of data.
- Serve as a test bed for the digital infrastructure called for in the IOM workshop (IOM, 2011).
Coalition Activities
The activities of the coalition could include:
- Convening the cancer community to seek consensus on opportunities and strategies for precompetitive data sharing.
- Advocating for adoption of standards for data collection, including appropriate content and use.
- Inviting and nurturing the participation of all information technology innovators to develop and sustain interoperable digital frameworks—composed of standards, specifications, vocabularies, and code bases—that enable users to exchange data through open interfaces among different repositories, institutions, or other sources of data. Importantly, such digital frameworks would not preclude or disrupt proprietary IT systems, as long as
open interfaces are maintained, to avoid the appearance or reality of undermining the commercial IT sector. - Serving as an honest broker within the biomedical ecosystem, linking those who need to use digital frameworks with those who provide them.
- Selecting and applying the standards and other infrastructure, including but not limited to those freely available from open-source and/or government informatics programs.
- Providing objective consulting services to ecosystem members seeking guidance on the best pathways to an efficient and effective digital infrastructure for data exchange.
- Providing project management services to ecosystem members who have a specific need for a digital framework, and who wish to have an honest broker oversee project implementation.
- Leveraging successful exemplars of research-enhancing data exchange to gain synergy by reusing and disseminating data-exchange capabilities across the cancer community nationally.
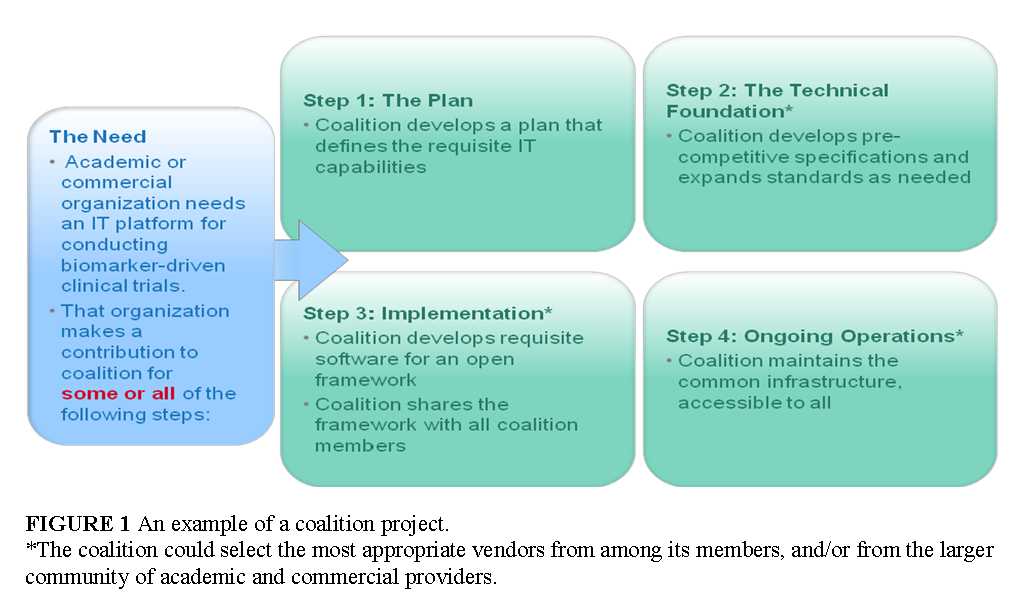
Participation
Any organization with a need for data exchange could bring a project to the coalition, and trigger some or all of the following activities: the development of a plan that defines the requisite IT capabilities; the development of precompetitive specifications and expanded standards, as needed; and the building and sustaining of the requisite IT infrastructure based on precompetitive specifications (Figure 1). The final outcome of such projects could be a common digital infrastructure accessible to all members of the stakeholder community.
Stakeholder Benefits
There are multiple ways that stakeholders across the entire continuum of cancer research and care could gain value from effective digital frameworks developed and used precompetitively to achieve data liquidity:
- Patient advocacy groups could share patient information with researchers to accelerate discovery of the molecular underpinnings of disease, and could link diverse biospecimen repositories interoperably to provide easier access and querying by researchers.
- Pharmaceutical researchers could gain the ability to interpret and share data that lead to the identification and validation of biomarkers of clinical significance, and to the identification and recruitment of subgroups that will respond to a marketed or experimental drug. Pharmaceutical companies could gain a national clinical trials infrastructure for multi-institutional trials that encompass imaging in order to develop their pipeline of new products. Pharmaceutical marketers could collaborate to share reports of adverse drug reactions (ADRs) on a global basis to identify molecular causes of ADRs within and across drug categories, potentially in collaboration with regulators.
- Clinical Research Organizations could gain a clinical trials infrastructure that leverages routine clinical care encounters to share data that are now wasted and must be regenerated multiple times in product development processes.
- Health care providers could share the clinical data about their patient populations with collaborators in order to widen their knowledge base, and adjust their standards of care to reflect new insights as they emerge.
- Software developers and systems integrators could gain products and services customized to the biomedical market. IT companies could benefit from the creation of a common market evolved from the fragmented components of the different sectors; from the reduction of risk that results from having a core collection of non-viral, open-source code that can be reused; and from the opportunity to leverage a substantial investment already made by the government in a common informatics infrastructure for cancer.
Funding/Sustainability
Numerous potential sources could be sought to help the coalition be self-sustaining, including
- Membership fees: The broad spectrum of stakeholders envisioned for this coalition may give it an advantage because larger entities are accustomed to paying larger dues. For example, the Personalized Medicine Coalition—an organization that serves as a “big tent” for many constituencies—has dues ranging from a low of $500 for patient advocacy groups to $25,000 for large corporations (Personalized Medicine Coalition, 2012). Comparable support for this coalition may potentially come from large pharmaceutical members as well as large IT companies that may see benefit in participation.
- Consulting fees: As described above, the coalition could be a first stop for those with a need for data exchange to determine potential solutions.
- Project management fees: The coalition could help ensure that those with a need for data exchange have the help of an honest broker throughout the life of a project.
- Nonprofit support: Other nonprofit organizations, such as foundations, may be focused on cancer as a national challenge as well as on the societal need for biomedical data exchange, and may provide funds for projects that serve as prototypes for national solutions.
- Government support: Within the Department of Health and Human Services, a multitude of projects are under way that need data-exchange capabilities; the coalition could compete for such grants and contracts on behalf of, or with, its members.
Activities of the coalition could be organized by volunteers at the beginning, and paid staff could be added as funds are raised over time.
Key Differentiators of the Coalition
In conceptualizing the new coalition, we have leveraged insights from various existing and previous models of collaboration in biomedicine, many of which were described at the IOM Precompetitive Collaboration Workshop in 2010 (IOM, 2010a).
Of greatest relevance are those collaborations designed to develop standards/tools; examples include nonprofit organizations such as the Pistoia Alliance (composed of life science companies, vendors, publishers, and academic groups that aim to create interoperability of research and development business processes) and SEMATECH (a partnership between the U.S. government and United States-based semiconductor manufacturers to solve common manufacturing problems) (IOM, 2010a).
No individual model, however, contains all the attributes that are required for success in achieving data liquidity. Therefore, the coalition envisioned here would be differentiated as follows:
- It would serve as a hybrid model in which individual entities could gain the benefit of data exchange and commercial IT companies could be remunerated, while still achieving the collective benefit of sector-wide data liquidity via open interfaces.
- It would seek to engage all the constituencies of the biomedical ecosystem, in contrast with models such as SEMATECH or Bellcore, which have involved primarily industry.
- It would not be a data aggregator or a central data warehouse; rather, it would be intended to facilitate data exchange via the pipes of common digital frameworks through which the data flow, with control remaining with the owner of the data. Thus, it would have the capacity to serve as an honest broker in addressing data-sharing challenges.
- It would augment the role of groups such as the American Society of Clinical Oncology and the American Association for Cancer Research in defining critical data to be collected by acting as an honest broker across all stakeholders.
- It would not be designed to benefit just one sector of the biomedical ecosystem, such as the pharmaceutical industry, in contrast with initiatives such as the Pistoia Alliance. Nor would it be designed to benefit just one geographic region, such as the Innovative Medicines Initiative (IOM, 2010a) in Europe. Nor would it focus on just one part of the biomedical continuum, such as the Partnership to Advance Clinical electronic Research
(PACeR, 2012). Its mission would encompass the informatics needs and benefits for all stakeholders along the spectrum of cancer and other disease research. - It would not compete with standard-setting bodies. Rather, it would be intended to capture and apply those standards that are already developed, and to catalyze the development of standards where none are available. Most importantly, it would attempt to disseminate both the standard and the software code to implement the standard. This approach is intended to be as pragmatic as possible and avoid perfection of standards that
gather dust on the shelf. - It would not be government-led, to ensure both full participation of all constituencies, and to acknowledge budgetary constraints in the current era. Rather, it would seek government participation and support in the form of contracts and grants, but would not expect or seek a dominant role for government.
- It would not undermine the role of the commercial IT sector, or seek to displace or disrupt proprietary software systems. Rather, it would require open interfaces between proprietary or open-source IT systems, to enable data liquidity while preserving intellectual property within the commercial sector.
References
- AACR (American Association for Cancer Research). 2011. Cancer progress report 2011: Transforming patient care through innovation. http://www.aacr.org/home/public–media/science-policy–government-affairs/cancer-progress-report.aspx (accessed February 6, 2012).
- B Corporation. 2012. Certified B Corporation. http://bcorporation.net/ (accessed February 6, 2012).
- Biomedical Research Integrated Domain Group. 2012. BRIDG. http://www.bridgmodel.org (accessed February 6, 2012).
- Bipartisan Policy Center Task Force on Delivery System Reform and Health IT. 2012. Transforming health care: The role of health IT. http://www.bipartisanpolicy.org/library/report/transforming-health-care-role-health-it (accessed February 6, 2012).
- Blumenthal, D. 2010. Launching HITECH. New England Journal of Medicine 362(5):382-385. https://doi.org/10.1056/NEJMp0912825
- Fenstermacher, D. A., R. M. Wenham, D. E. Rollison, and W. S. Dalton. 2011. Implementing personalized medicine in a cancer center. Cancer Journal 17(6):528-536. https://doi.org/10.1097/PPO.0b013e318238216e.
- I-SPY 2. 2012. I-SPY 2 TRIAL. http://ispy2.org/ (accessed February 16, 2012).
- Internet Engineering Task Force. 2012. The tao of IETF: A novice’s guide to the Internet Engineering Task Force. http://www.ietf.org/tao.html (accessed February 6, 2012).
- IOM (Institute of Medicine). 2009. Beyond the HIPAA Privacy Rule: Enhancing Privacy, Improving Health Through Research. Washington, DC: The National Academies Press. https://doi.org/10.17226/12458.
- IOM. 2010a. Extending the Spectrum of Precompetitive Collaboration in Oncology Research: Workshop Summary. Washington, DC: The National Academies Press. https://doi.org/10.17226/12930.
- IOM. 2010b. A Foundation for Evidence-Driven Practice: A Rapid Learning System for Cancer Care: Workshop Summary. Washington, DC: The National Academies Press. https://doi.org/10.17226/12868.
- IOM. 2010c.A National Cancer Clinical Trials System for the 21st Century: Reinvigorating the NCI Cooperative Group Program. Washington, DC: The National Academies Press. https://doi.org/10.17226/12879.
- IOM. 2011. Digital Infrastructure for the Learning Health System: The Foundation for Continuous Improvement in Health and Health Care: Workshop Series Summary. Washington, DC: The National Academies Press. https://doi.org/10.17226/12912.
- IOM. 2012a. Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington, DC: The National Academies Press. https://doi.org/10.17226/13297.
- IOM. 2012b. Facilitating Collaborations to Develop Combination Investigational Cancer Therapies: Workshop Summary. Washington, DC: The National Academies Press. https://doi.org/10.17226/13262.
- IOM. 2012c. Informatics Needs and Challenges in Cancer Research: Workshop Summary. Washington, DC: The National Academies Press. https://doi.org/10.17226/13425.
- IOM, and NRC (National Research Council). 2006. From Cancer Patient to Cancer Survivor: Lost in Transition. Washington, DC: The National Academies Press. https://doi.org/10.17226/11468.
- Loten, A. 2012. With new law, profits take a back seat. Wall Street Journal, January 19. http://online.wsj.com/article/SB10001424052970203735304577168591470161630.html (accessed February 6, 2012).
- NCI (National Cancer Institute). 2011. An assessment of the impact of the NCI Cancer Biomedical Informatics Grid (caBIG®): Report of the Board of Scientific Advisors Ad Hoc Working Group. Bethesda, MD: NCI.
- NRC. 2011. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: The National Academies Press. https://doi.org/10.17226/13284.
- PACeR. 2012. The Partnership to Advance Clinical electronic Research (PACeR). http://www.pacerhealth.org/ (accessed February 6, 2012).
- PCAST. 2010. Report to the President: Realizing the full potential of health information technology to improve healthcare for all Americans: The path forward. http://www.whitehouse.gov/sites/default/files/microsites/ostp/pcast-health-it-report.pdf (accessed February 6, 2012).
- Personalized Medicine Coalition. 2012. Personalized Medicine Coalition. http://www.personalizedmedicinecoalition.org/ (accessed February 6, 2012).
- The McKinsey Global Institute. 2011. Big data: The next frontier for innovation, competition, and productivity. http://www.mckinsey.com/Insights/MGI/Research/Technology_and_Innovation/Big_data_The_next_frontier_for_innovation (accessed February 6, 2012).