A Proposed Approach for Implementing Genomics-Based Screening Programs for Healthy Adults
The central idea of early disease detection and treatment is essentially simple. However, the path to its successful achievement (on the one hand, bringing to treatment those with previously undetected disease, and, on the other, avoiding harm to those persons not in need of treatment) is far from simple, though sometimes it may appear deceptively easy. – Wilson and Jungner, Principles and Practice of Screening for Disease [1]
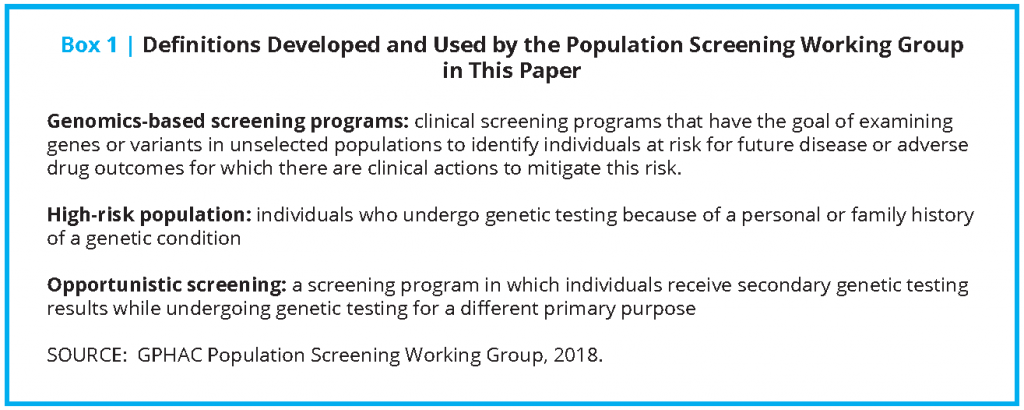
Introduction
In 2017, the Genomics and Population Health Action Collaborative (GPHAC), an ad hoc activity of the Roundtable on Genomics and Precision Health of the National Academies of Sciences, Engineering, and Medicine, formed the Population Screening working group (PSWG), focused on genomics-based screening programs in healthy adults. This group identified the various aspects of implementation that need to be considered when developing a large-scale, targeted sequencing program. Evidence indicates that 1 to 2 percent of the overall US population has a pathogenic variant putting them at a substantially elevated risk of a serious yet preventable disease [2,3]. There are well-established health interventions for individuals identified to have a pathogenic variant through a personal and/or family history of such conditions (high-risk populations). As a result, there is an opportunity to use screening to identify these otherwise healthy individuals and consider implementing such interventions, thereby reducing the risk of undesirable outcomes caused by missed diagnoses. However, there are also important issues for the field to address as these programs develop, including feasibility of screening, potential benefits and harms, outcomes, costs, and ultimately, clinical utility.
The following potential roadmap was developed by participants in the GPHAC Population Screening working group through a series of facilitated discussions, as a suggested starting point and educational resource for groups planning to carry out projects involving genomics-based screening programs in the healthy adult population. The roadmap will likely need to be updated as new evidence and technology becomes available. Indeed, it became evident through discussions with this group that much data are needed before we know whether routine implementation of genomic screening in the population is warranted. As Wilson and Jungner remind us in the epigraph above, implementing screening is fraught with potential complications. It is crucial that all efforts to pilot such an approach include robust methods for subsequent data collection, including outcomes, benefits, harms, and costs to lay a foundation for evidence-based realization of the promise of genomics-based screening programs.
This paper will cover the optimal genes to include in genomics-based screening programs, the complexity of identifying a point in time at which to begin screening, who should perform the screening, where the programs should be administered, along with ethical and economic considerations.
Optimal Genes for Genomics-Based Screening Programs
After much deliberation, participants in the PSWG decided that the first step in developing considerations for those organizations interested in developing genomics-based screening programs is to suggest the most appropriate genes for possible screening and develop a rationale for their use in such a pilot program. Given the clear evidence of our current inability to reliably interpret the results of genome-scale sequencing in the general population [4], the PSWG chose to focus on a more limited public health goal, namely the prospect of targeting well-studied genes (and well-characterized alleles within them) linked to conditions with well-studied, effective clinical interventions for purposes of primary screening in the general population. Such an approach will allow for provisionally implementing and investigating a targeted version of genomic screening that stands the best chance of offering a high ratio of benefit to harm in the general population.
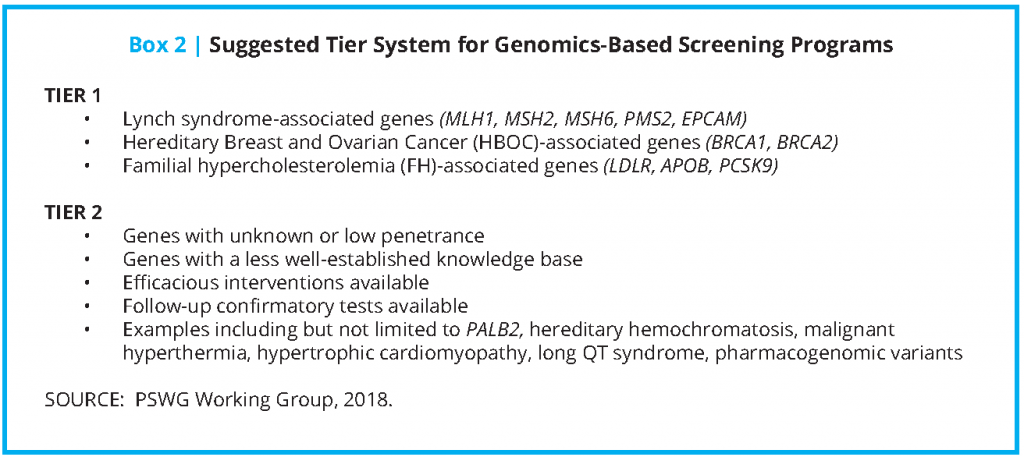
In developing a list of potential genes for population screening, the group considered the existing difficulties inherent in variant assessment, the risk of false positive results, and the potential for over-diagnosis given the low prior probability that those screened actually have a disease. With this in mind, the existing recommendations from the American College of Medical Genetics and Genomics (ACMG) for opportunistic screening of existing data were considered [5,6]. Although the ACMG’s list of 59 genes does include gene-condition pairs where there is evidence for use of screening to drive overt recommendations for beneficial medical intervention, there is less supporting evidence for other gene-condition pairs on the ACMG list. The PSWG decided that 10 of the 59 genes, namely those associated with the three conditions in the CDC’s Office of Public Health Genomics (OPHG) Tier 1 Genomics Applications, were fully in line with the screening priorities of the PSWG [7].
The working group developed their own two-tiered approach, and ultimately settled on a limited set of genes that could fit within the following screening categories (that will be referenced throughout the proposed roadmap):
- “Tier 1” consists of those genes with high penetrance (the probability that disease will appear when a disease-related genotype is present), well-understood links to disease, and well-established, effective interventions that result in substantial prevention or mitigation of disease or disease risk.
- “Tier 2” consists of those genes that could be possible candidates for screening but where there may be disagreement about whether there is sufficient evidence for population screening. The authors believe that some groups who wish to carry out pilot screening programs may find certain genes compelling, whereas others may not consider them to be good candidates for screening. Examples include genes with certain characteristics such as low penetrance (e.g., HFE mutations associated with hereditary hemochromatosis) and/or a less complete knowledge base (e.g., PALB2 mutations associated with certain types of cancer). Although there are no definitive and universally agreed-upon criteria that qualify a gene as a candidate for screening, it should be emphasized that a strong justification regarding specific actionability should be present. Tier 2 genes should be “close calls.”
The PSWG acknowledged that there could be circumstances where a pilot population screening study may choose to include certain Tier 2 genes based on features of the study population, expertise within the study team, availability of high-quality secondary screening tests, advances in the knowledge base associated with the gene-condition pair, or other valid considerations. In such cases it will be especially important to have clear discussions with study participants about the limitations of current knowledge and to collect even more extensive follow-up data (e.g., clinical outcomes, penetrance) given the increased, inherent uncertainties of those genes.
In summary, although there are a number of genes that could be considered for a pilot program, the working group focused on a set of three criteria in developing their Tier 1 list:
- genes that, when disrupted by a pathogenic variant, are associated with a high risk of developing a disorder associated with serious morbidity and/or mortality;
- well-established, acceptable, highly effective, and specific preventive interventions for those identified as at risk; and
- an established knowledge base regarding the gene and the condition(s) associated with pathogenic and likely pathogenic variants in the gene.
The working group also discussed what genes and variants would be in-scope and out-of-scope for population screening, and these examples will change over time as the evidence changes. Genome-wide association study findings and risk-raising single nucleotide polymorphisms, though a rich source of data, were excluded given the wide breadth of relative risk, lack of established clinical usefulness, and evidence limitations thus far, as were nonmedical traits and conditions with no available treatments (e.g., early-onset Mendelian Alzheimer’s disease). In terms of reporting results to the patient, the PSWG considered known and likely pathogenic variants to be in-scope and variants of unknown significance (VUS) to be out-of-scope in this context. All variants should be systematically adjudicated by well-accepted and rigorous criteria [8]. Although the decision to exclude VUS findings is consistent with ACMG recommendations regarding the return of secondary findings, there are limitations with this approach worth considering that are outside the scope of this paper. For example, some VUS results will ultimately be reclassified as pathogenic as more evidence becomes available. In the future, screening programs may wish to consider mechanisms for reevaluating these variants and recontacting participants.
Considering the Right Age for Screening
Screening is looking for disease in those who do not have symptoms. In general, there are two types of screening. The first type is done in the general population, and the second type is done in those who are at high risk for a particular condition. One example of general population screening is phenylketonuria (PKU) screening in newborns, performed because there are immediate interventions available to prevent intellectual disability in newborns with the condition and because amelioration of the condition is not possible once symptoms manifest. Additional examples of general population screening include screening for high blood pressure in adults aged 18 and older and colorectal cancer screening in adults. High-risk screening in people with a personal and/or family history of disease, or a known underlying genetic predisposition, differs in intensity and/or starting age compared with screening in the general population.
The age at which to start screening is determined by many factors, including the prevalence of the disease, its natural history, the sensitivity and specificity of the test, and the effectiveness of the treatment. Although genetic testing offers some advantages (e.g., no need to consider repeat testing in most cases), the young and evolving field is challenged with evidence gaps, education deficits, and implementation issues, among others.
Each Tier 1 condition is unique with respect to the ideal age for therapeutic intervention for those who carry pathogenic variants. Although the genetic results do not change with age, it is likely that a single age for all testing is not optimal. For example, there is evidence that lipid-based testing for FH between the ages of 9 and 11 is beneficial, because early interventions can affect outcomes. However no DNA-based population screening recommendations for pediatric FH are currently in place [9]. In the case of HBOC, the National Comprehensive Cancer Network does not recommend initiating breast MRI screening until the age of 25. Despite this recommendation, the optimal age for HBOC still remains unclear. It has been recommended that 30 years of age may be the right time to screen women since few women will have had a cancer by that age [10]. Others argue that by 30 years of age some at-risk women will have developed cancer and many women will have had children and would have wanted to know about their BRCA status before conceiving.
The issue is made more complicated because screening is based on the assumption that the penetrance of a mutation in the general population is the same as the penetrance in a high-risk population, but this is not necessarily true. Thus, although we may know an individual’s risk variant after screening, we do not necessarily know the individual’s specific disease risk conferred by the test result. This makes accurate genetic counseling and use of those test results problematic for the patient and their providers, and could result in undertreatment (e.g., insufficient management) or overtreatment (e.g., unnecessary risk-reducing surgeries).
All of this means that the nascent nature of the field should be acknowledged and care should be taken moving forward. Although it is possible that in the future, as the number of Tier 1 conditions increases, “packaged” screening for more than one disease at a time will gain ascendance, for now, the decision of when to screen the general population is likely to be different for each indication until further data are forthcoming. Finally, cautious implementation of screening programs in the general population should include careful, formal evaluation of the intended consequences, including outcomes.
Who Should Perform Genomics-Based Screening?
Screening the general population for the presence of Tier 1 genetic variants offers the opportunity to save lives and prevent disease; however, several important issues should be considered before implementation. The first issue is the absence of guidance from professional organizations and medical societies on when genomic screening in the general (i.e., average-risk) population is appropriate. This is not surprising, given the lack of demonstrable clinical utility to date. The development of guidelines on genomic screening in the healthy adult population, clinicians and health systems, and ready cost coverage by payers for services related to genomic screening and follow-up will be contingent on pilot studies and data aggregation that demonstrate a sufficient evidence base. Limiting initial screening to a panel of Tier 1 genes may make the most sense in terms of the programmatic costs for well-designed pilots.
The Role of Public Health in Genomics-Based Screening
State newborn screening (NBS) programs have existed since the 1960s, and NBS is often considered to be one of public health’s most singular achievements [11]. Most states indicate high levels of compliance with NBS programs, often as high as 99.9 percent [12]. However, such extraordinary compliance is unique and largely a reflection of the fact that NBS is considered so essential to the public good that “opt-in” consent is deemed unnecessary. These factors do not exist for any other screening program and will not exist for genomic screening of adults likely because of logistics, feasibility, lack of evidence for clinical utility, and resources. Thus, genomic screening of adults will require new mechanisms for collecting samples and obtaining consent, along with ensuring appropriate follow-up services.
It is likely that genomics-based screening could be implemented in the health care delivery sector much like population-based screening for aneuploidy in pregnant women or breast cancer and colorectal cancer screening of adults via mammography and colonoscopy. When stakeholders believe there is sufficient evidence to support screening, as is the case with the aforementioned examples, then offering such screening becomes standard of care for clinical practice and is carried out in clinicians’ offices or other health care settings [13]. It is worth mentioning that rates of patient compliance with adult health screening procedures such as mammography and colonoscopy tend to be far lower than NBS programs.
Even if genomics-based screening takes place in the health care delivery setting, there is a role for the public health sector. Public health practitioners routinely work to implement validated applications with proven health benefits and evaluate their effects on health care or the health care delivery system [13]. The CDC’s core public health functions and 10 essential services (Figure 1) create the framework for all public health services, including public health genetics/genomics [14]. Collaborative approaches between health care organizations and the public health sector will have highly desirable synergies.
Possible Settings for Genomics-Based Screening Programs
When developing a genomics-based screening pilot program, groups should consider the many options that are available for reaching a wide audience. Some potential settings for engaging populations and carrying out genomics-based screening programs are described below along with the rationale for and against each scenario. This is not a comprehensive list, and other models (e.g., nonstaff model HMOs) should also be considered.
Payer-Provider (Integrated) Health Systems
Integrated health systems, specifically those that have an attached health insurance plan, offer considerable opportunities for implementing genomics-based screening programs. Because of the connections between providers and reimbursement systems in these networks, there may potentially be fewer obstacles regarding the decision-making process to implement as well as the requirements for reimbursing those services. While there is variation among integrated systems, they are broadly defined as “an organized, coordinated, and collaborative network that links various healthcare providers to provide a coordinated, vertical continuum of services to a particular patient population or community” [15]. Those systems are also financially accountable and incentivized to work toward improved outcomes in their patient populations.
Certain advantages inherent to integrated systems may provide a good rationale for using them as a setting to implement genomic screening programs. The focus placed on health outcomes is one. For instance, Gillies et al. found that integrated systems performed at a higher level in terms of outcomes in preventive measures like heart disease and women’s health screenings compared with nonintegrated systems [16]. If genomic screening programs were shown to improve outcomes among the patient population, the coordinated nature of integrated systems could encourage a pay-for-performance model of reimbursement and reduce some of the economic challenges of implementation. Other rationales include access to a broader and more diverse population in terms of socioeconomic status and ethnicity as well as increased engagement opportunities with patient populations. Finally, there is less fragmentation of care and clear opportunities for standardizing follow-up care for patients within such systems since patients typically have all their medical needs filled through a single administrative organization. This reduces some of the complexities of implementing risk-reducing modalities and tracking outcomes.
On the other hand, integrated systems may also have disadvantages. One is location and the associated logistical challenges that arise when trying to coordinate care across a large geographical area. For some integrated systems, including smaller systems or those located in rural areas, genetics expertise may not be readily available. Although one could imagine that challenges related to expertise are not unique to implementing genomic screening in this setting, a literature review by Armitage et al. found that some integrated systems had staff-related challenges when introducing new skills or knowledge areas for new providers [17]. Finally, although integrated systems may have access to a broader and more diverse patient population, some individuals without coverage may also be excluded, therefore exacerbating disparities.
University-Associated or Research Hospitals
Forming an alliance with a university-affiliated or research hospital to offer genomics-based screening may encourage more widespread implementation and better data collection than certain other models. One example of such an approach is the New England Newborn Screening Program, which has operated under the auspices of the University of Massachusetts Medical School since 1997. The New England Newborn Screening Program serves families in five states and provides high-quality, low-cost laboratory screening, clinical follow-up, and research to prevent or minimize the effects of serious genetic conditions. One advantage of this type of approach is that the university staff who carry out the testing and analysis have a very high comfort level with laboratory technology, collection and analysis protocols, and steps for follow-up counseling and treatment. Research or university-associated hospitals are often well prepared to handle the logistics of a large-scale genomic testing program, and their geography can be an advantage because they are able to reach a large swath of the local population.
However, there are limitations to offering screening through a university-associated or research hospital. One such challenge is that other competing research interests may make it difficult to secure funding for this type of program. Funding that comes from short-term grants may make it challenging to sustain a program after the grant term has ended. Also, university-associated research hospitals are often in urban settings, which would potentially exclude rural populations from participation. Finally, care is often fragmented in such settings, making follow-up potentially less optimal.
Self-Insured Employers
A third arena in which to carry out a genomics-based screening program is through a self-insured employer’s health benefit program. Heart disease and cancer are the two leading causes of death in the United States [18], and self-insured employers have started offering genetic testing for certain clinically actionable conditions as part of an overall preventive health strategy. There are several incentives for employers to consider this approach, including the desire to detect cancer and heart disease earlier. Furthermore, some employers believe that offering health perks such as access to genetic testing is an effective way to engage with employees and increase their overall job satisfaction. Another benefit to carrying out genomics-based screening through self-insured employers is that all participants have access to insurance coverage for any necessary follow-up testing and preventive interventions.
Genomics-based screening programs offered through self-insured employers present some possible disadvantages as well. Certain benefits to the company, such as long-term savings in health care costs, may be realized only if employees remain at the company for a certain period of time after the genetic screening. In addition, care would need to be taken to ensure that employees feel confident in their freedom to decline to participate. Concerns about the privacy of health information may be relevant, given that individuals may be worried that their genetic test results will be shared with their employers. Protecting the privacy of employees and their health data is of the utmost importance, and personally identifiable health information of participants should not be shared with employers unless there is proper consent to the disclosure of such information.
Generalizability and scalability of such efforts are also a potential disadvantage since, by definition, participants must be employed by participating businesses. Although large corporations that can afford to facilitate such a program often employ people from a range of socioeconomic classes, screening may be cost prohibitive for smaller companies, so testing may reach only a limited audience (e.g., employees of a large corporation). Thus, there is a concern that rolling out screening in such a setting could lead to significant weighting of those screened toward those who are employed by a large corporation.
Military
Engaging the military around genomics-based screening is another option to consider. One reason this may work well is that the military is already engaged in genetic testing. For example, the US Air Force has a Medical Genetics Center, which serves as the reference genetics lab for the entire Department of Defense, as well as its own Personalized Medicine Program that coordinates a variety of genomics-focused projects [19].
This population, including service members and veterans, may be motivated to volunteer for opportunities such as population screening, in which they can contribute to both research and the improvement of their own health and the health of their families. Leaders of one genetic testing pilot project in the Air Force (MilSeq) indicate that service members have not expressed fears about the misuse of genetic information. Those who have declined tend to do so because of a general disinterest in joining a research project.
Another unique advantage to including the military is its meticulous collection and storage of health care data of all active-duty service members and veterans, and interoperability of that health data as they move throughout the military health system. This health information follows them, their families, and survivors throughout their military career and retirement (if they choose to stay covered by TriCare and then the Veterans Administration). In a health care environment focused heavily on value, feeding targeted and actionable genomics data into an already rich, integrated, and contained data source can provide potentially helpful health and economic outcomes.
There may be unique challenges associated with engaging the military, particularly related to genetic discrimination. Members are not covered under the Genetic Information Nondiscrimination Act (GINA), although several similar policies exist and apply to the military population. Although previous policies stipulated that service members would be denied benefits if they developed a condition (including but not limited to genetic) during their time of service that requires discharge, the 2008 National Department of Defense Act clarified that this is the case only if symptoms of the condition were present at enrollment. In other words, a diagnosis based on a genetic test result alone does not constitute a preexisting condition. Specific branches of the military may have additional policies and/or protections, however. For example, Air Force Instruction 36-2706 explicitly prohibits discrimination on the basis of genetic information. However, the military does have a history of using genetic information to restrict an individual’s participation in physically demanding tasks (e.g., screening for hemoglobinopathies and glucose 6-phosphate dehydrogenase deficiency) [20]. Supervisors may be particularly interested in screening results for Tier 2 cardiovascular conditions such as long QT syndrome, which could potentially be used to preclude a service member from participating in certain missions. It will be important for any genomics-based screening program to carefully consider the existing protections and practices surrounding both employment and insurance discrimination, and provide informed consent to participants accordingly. Finally, the selective (all recruits and active-duty members are evaluated for fitness) and self-selective nature of the military may lead to representation challenges.
[NOTE: The authors of this paper focused this section on genetics and genomics programs available for military personnel. It is also important to note that the Department of Veterans Affairs’ (VA) Office of Research and Development houses a genomic research program covering genomic and phenotypic studies of recruited veterans into the Million Veteran Program (https://www.research.va.gov/mvp/). The VA has surveyed veterans and found support for genomic testing and the use of these data with medical record information for large-scale interpretation, research, and analysis. With regard to protection under the Genetic Information Nondiscrimination Act (GINA), VA benefits are not denied based upon a newly uncovered germline genetic predisposition. Rather, it is assumed that veterans were considered in good health upon conscription into the Armed Forces. Secondary illnesses, including those with germline predisposition, would not deter benefits for the veteran if not present at time of conscription.]
Informed Consent and Access to Genetic Counseling
Regardless of the model(s) employed for implementation of pilot programs, access to expert genetic counseling before and after the delivery of test results to participants is critical. Entities implementing genomics-based screening programs must ensure that participants have access to genetic counseling from trained professionals, so that participants can get sufficient and accurate information about the test results that allows them to use that information to make decisions about future medical care. Understanding the implications of positive results implies appropriate follow-up so that effective prevention can be implemented. For example, those found to have a pathogenic variant in a Lynch syndrome-associated gene need referral for frequent colonoscopies, those with a BRCA1/2 pathogenic variant need referral to a high-risk clinic that can educate and implement enhanced surveillance or risk-reducing surgery, and those found to have familial hypercholesterolemia will require follow-up by a primary care provider or cardiologist for appropriately aggressive lipid management. Adequate education and resources must also be in place for those who test negative, because it is critical for participants to understand the limitations of a negative result within a targeted screening approach. For example, negative BRCA1/2 or Lynch syndrome testing does not exclude the possibility of other hereditary cancer syndromes.
It will likely be necessary to use creative service delivery models to address the limited workforce of genetics professionals. Buchanan, Rahm, and Williams describe several alternate service delivery models for genetic counseling in their recent review, including group counseling, nongenetics clinician counseling, and pre-and post-test counseling [21]. The authors of this paper would emphasize that in a broad population screening context it will not be necessary for all individuals to have contact with a genetic counselor. Indeed, most screened individuals will likely not need traditional counseling. However, access to professional counseling is critical for those individuals with questions or unusual circumstances (e.g., a strong family history of the condition being screened).
Population screening is a departure from the traditional approach of high-risk population testing, in which individuals typically receive pre-test genetic counseling that includes an in-depth discussion of the benefits and limitations of testing, with particular attention to the psychological and social effect that testing could have on both an individual and a family. After these sessions, some individuals ultimately decide not to proceed with genetic testing for a variety of reasons. It will be important for systems to consider the resources available for providing pre-test counseling in population screening programs. Such counseling may be provided by other models rather than traditional genetic counseling (e.g., using prepared literature, videos, or web interfaces). Among other concerns, participants should be aware of the limitations of GINA, which currently does not protect against discrimination for long-term care, life, or disability insurance. Again, all of this is to say that systems should proceed with caution when implementing screening programs, ensuring that recruitment strategies adequately address issues of informed consent.
Engagement Strategies
If an institution decides to implement a genomics-based screening program, a strategy for raising awareness and encouraging participation should also be developed and measured. Similarly, if an institution is developing a pilot study or returning results about Tier 1 genomic conditions as part of an ongoing research program, then strategies for both recruitment and longitudinal engagement need to be developed and measured. The strategies for awareness and participation are similar to strategies for recruitment, whereas longitudinal engagement requires separate considerations.
For health systems, researchers, and employers with on-site clinics, large-scale participant recruitment can be optimized through physical integration with on-site clinics. For example, Geisinger’s MyCode initiative uses full-time consenters working with patients in outpatient clinic settings to inform them of the program and enroll them in this research initiative with consenting success rates in the 80–90 percent range [22]. Other recruitment strategies include recruitment through primary care physicians, advocacy groups, referral from a past participant, and posters. In addition, structured educational events held at the institution can be tied to consent and/or sample collection. A growing body of literature supports digital and social media’s effectiveness in recruiting study participants, including historically hard-to-reach populations [23,24]. Although consumer genetics companies such as 23andMe have had success in using digital and social media to recruit participants and collect longitudinal health data, the target audience for screening initiatives is not well defined, and without a defined denominator the recruitment success rate is unclear.
Genomics-based screening is just the first step in early detection and prevention of Tier 1 genomic conditions. Ongoing engagement of participants is necessary to ensure compliance with risk-appropriate surveillance/prevention guidelines as well as to assist in collecting necessary longitudinal research data. Health systems should consider implementing downstream research studies in conjunction with genomics-based screening programs to fill the evidence gap in clinical utility. The All of Us research program, an effort to collect data from one million participants in the United States to accelerate research and improve health, has made it a priority to communicate and engage with Americans from all communities, so there may be lessons and best practices to take away from that effort. Engagement is particularly important for health systems that would like to conduct research, because it enables further phenotypic, lifestyle, or wearables data to be collected longitudinally. A number of digital strategies can be deployed to increase both engagement and compliance:
- Return of results—If participants have given their consent, regular return of information from the study organizers can offer many potential benefits, including increased recruitment and retention. Investigators should routinely examine whether and how to return individual results on a study-specific basis. Such information may include ongoing aggregate data regarding participation, results, and publications. The time and process through which results will be communicated should be addressed in the consent process.
- Use of mobile devices and secure internet EHR portals—Mobile device apps and EHR portals can be used to deliver educational disease-centric content as well as create reminders for annual screenings and medication.
- Family tree/history tools—Enabling participants to build their family tree digitally and keep their own records of their family health history can serve two functions: tracking how a trait or condition can be passed down between generations, and identifying other at-risk family members. Family tree tools that can be shared with family members may also serve to increase awareness of population-based screening programs and encourage others to participate.
Economic Considerations
As genomics-based screening programs are introduced into clinical settings to improve population health, the economics of the clinical value and utility of genomic information become critical in decision making. The types of economic evaluation conducted depends on several factors: (1) the chosen perspectives of the decision makers (e.g., societal, patient, third-party payers, health care system); (2) the outcomes of interest (e.g., life years saved, quality-adjusted life years saved); (3) cost data (medical versus nonmedical cost); and (4) the comparators. The value of comparison to an existing practice of health care services or the alternative intervention can help determine the added costs and health benefits the new intervention provides over and above the existing ones. Modeling can be used to compare the expected value and cost across different strategies (e.g., status quo standard of care versus genomics-based testing diagnosis or comparing two new alternatives) to provide decision makers with important information.
Although any economic model may be complex, there are special considerations in genomics-based screening programs. The counting of the economic and health costs for any screening program is profoundly influenced by the time horizon (how far into the future the effect of the outcomes extend). However, when dealing with genetics, a well-described risk to other family members and future generations is often significant. Consideration must be given to whether and how to count economic and health outcomes for future generations and for current family members. This also influences where to get the outcome data, since any future economic and health costs may change with new treatments. Although this also provides challenges for creating the model, economic modeling can help inform recommendations for health insurance coverage, evaluate the optimal strategy for adoption, and assess the level of uncertainty in implementing genomic testing in public health programs. This can be used to compare the expected value and cost across different strategies to provide policy makers with important information.
Cascade Testing of At-Risk Family Members
Within genetic testing, some conditions included in the list of Tier 1 conditions may also suggest a potential benefit in using cascade screening to identify at-risk family members. Each first-degree relative of a proband (the first individual in a family found to be affected with a genetic disorder) with an autosomal dominant condition has a 50 percent chance of carrying the same mutation (not considering de novo variation). One goal of cascade testing is to identify at-risk relatives who require careful screening from those who do not. Once a proband is identified, a concerted effort should be made to make testing available to their at-risk family members. However, there can be significant barriers to family communication about genetic risk information, including psychological, educational, and geographical challenges. It is important to consider family dynamics and ethical factors, including preserving confidentiality and privacy, minimizing psychological harm and genetic discrimination, and balancing the right “not to know” with “duty to warn,” among others [25].
In her paper cited above, Sturm also discussed several challenges in cardiovascular cascade testing that may be relevant for other conditions, including barriers to reimbursement for genetic counseling services, insufficient knowledge about genetic risk assessment by health care providers, a shortage of genetic counselors (especially in rural areas), and an individual approach to disease prevention as opposed to a family-centered design [25]. Although traditional methods of communication usually involve communication among family members and/or clinicians, screening entities can provide assistance. For example, when individuals test positive for a pathogenic variant, they can inform first-degree relatives by using a letter provided by the testing entity that is written in simple terms. In addition, commercial genetic testing laboratories have developed web-based programs that facilitate lower-cost testing for at-risk relatives. These programs are designed to reach across health systems, states, and countries in a regulatory compliant manner. The GPHAC Cascade Screening working group is identifying some of the potential opportunities for implementing cascade screening more widely and plans to develop tools and publications highlighting them [26].
Ethical Considerations for Genomics-Based Screening Programs
Ethical considerations should loom large when contemplating large-scale population screening. The ethics of genomics-based population screening can be viewed from both clinical and public health perspectives. Therefore, developing such screening programs demands a hybrid clinical–public health ethical approach.
Clinical ethics tends to focus on individual autonomy and issues related to respect for people, including privacy, consent, and reporting requirements. By contrast, public health aims to promote and maximize population health, often through systemic measures and policies that adopt a utilitarian approach that weighs opportunities and benefits against costs and harms and attempts to maximize beneficial outcomes for the majority of the population. On the issue of justice, clinical ethics focuses on individual access, whereas public health tends to focus on how to fairly distribute social goods among a group. Table 1 below contrasts the perspectives on ethics from a clinical standpoint versus a public health standpoint.
Whereas autonomy in clinical ethics often trumps other considerations, many issues in public health arise as a result of conflicts between personal liberty and the common good. Examples where the common good takes precedence include mandating vaccines for school entry, restricting smoking in public buildings, and barring people from flying in commercial aircraft if they have a highly infectious disease such as multi-drug-resistant tuberculosis.
Can an analogous public health ethics rationale be provided for the case of genomics-based screening? For example, consider a high-risk group for a particular genetic condition that can be prevented by timely screening and treatment. Do those individuals have a duty to be screened? What if waiting for symptoms to manifest would raise treatment costs over a lifetime? Their individual decision not to screen could burden the health system and indirectly harm everyone else by diverting resources from other, more urgent conditions to what would have been an easily preventable condition. The reasoning here is related to why insurance companies have higher premiums for smokers; namely, they indirectly raise costs on the system for everyone, including nonsmokers.
Ethically speaking, these remain contentious issues. Further debate and discussion involving multiple stakeholders, as well as robust outcomes data and cost-benefit analyses of screening, will be important in helping society make decisions around implementation of genomic screening.
If, as outlined above, the goal of genomic screening of populations is to help maximize public health, then one can argue that whether to screen at all, and what to screen for, are salient ethical issues, because each commitment to finding a Tier 1 genetic predisposition presents both an opportunity and a cost. In the context of newborn screening (NBS), the Wilson-Jungner criteria have provided a guide for what conditions to include on state-mandated screening panels for more than five decades [1]. Examples of criteria for the inclusion of certain diseases include significant morbidity and mortality and the availability of effective treatment. However, the process of selecting which conditions to include on NBS panels is not straightforward. Moreover, although testing for additional conditions may only minimally increase the up-front cost of an NBS test, there is the risk of unforeseen harms through false positives (in the case of rare conditions) or the discovery of conditions with variable presentations, ages of onset, and/or insufficient adverse outcome data. Similar complications pertain to the consideration of population-based screening for adult-onset diseases: what constitutes an “actionable” genetic predisposition, and who gets to decide? How often will “the list” of those screened for genes and/or diseases be revisited, and by whom? What steps will be taken to ensure the validity of the information communicated to patients?
Once the decision to screen for particular causative alleles has been made, there is an ethical imperative for those whose health is at greatest risk to be ensured access to available screening at minimal financial cost to them. In the context of screening under the auspices of self-insured employers or integrated health systems with attached insurance plans, complications arise, since most individuals are not employed by the same entity for long periods of time. Likewise, in the fragmented health system of the United States, where access to health care is dependent upon employment and political vagaries, ensuring access to needed preventive modalities looms as a major problem confronting the success of genomic (and all other) public health efforts.
NBS is instructive here as well: although it is typically performed without regard to insurance status or parents’ ability to pay, most states collect a fee for it [27]. In some cases, this fee is covered by the patient’s insurance, but for low-income families it is often covered by the State Children’s Health Insurance Program or Medicaid. Of note, conditions on the Recommended Uniform Screening Panel are considered preventive services that certain insurance companies are required to provide coverage for under section 2713 of the Public Health Service Act. Preemptive protections for individuals being screened should be considered and discussed.
Delivering and paying for genomics-based screening becomes even more complicated in ways that NBS is usually not when one considers the likelihood that any predisposition that is identified in the screening will be shared with family members and hence suggest the need for identification and cascade screening of first-degree and possibly more distant biological relatives. Given that any such genomic program will find a nontrivial number of Tier 1 alleles in the population [28], screeners will be obligated to develop a plan to assist the positive-result recipient with informing family members about potential risks to them (as well as caveats about false-positive and false-negative findings) [29]. Again, this will require planning to accommodate for the costs, tools, and other resources necessary for additional screening, counseling, and follow-up referrals for treatment [30].
Questions of privacy, confidentiality, and access to data are intrinsic to any genetic screening or testing program. One obvious and reflexive approach to mitigating these is to embed them in informed consent documents. Unfortunately, consent forms are often poorly understood [31] and poorly recalled [32]; they can often appear to be exercises in liability mitigation rather than clear explanations of experimental procedures designed with the patient or research participant in mind [33, 34, 35]. Efforts to optimize consent for genomics-based screening programs should be pursued, particularly if included alongside other routine health screenings. Ideally, any large-scale screening program would initiate community consultation well before patient/participant recruitment. Investigators might also consider remote, video-based (or other visual), self-guided consent processes [36]. Such processes should also include a frank discussion of the limitations of our current knowledge and management recommendations, as while as information about what happens in the event of a data breach.
Conclusion
The advent of massively parallel sequencing has already transformed the clinical practice of medical genetics, serving as an indispensable tool for the diagnosis of diseases that have a primarily genetic etiology. The ability to readily examine panels of selected genes or entire exomes or genomes is a valuable adjunct to the diagnostic process, but currently remains directly applicable to a relatively narrow range of patients—that is, those with Mendelian disorders.
How we might use this same technology to benefit the general population is one of the most exciting tasks now confronting the field of genomics. The potential gains inherent in worthwhile public health interventions are enormous, in part simply because of the high number of those receiving the intervention. However, for the same reason, public health interventions carry significant potential risks, and examples of screening modalities that have stood the test of time and have been demonstrated to be worthwhile are limited. Unlike in the clinical setting, public health applications are implemented en masse. Those on the receiving end of public health interventions start from a position of apparent health and may never fall ill from the targeted disorder, regardless of a particular intervention. Moreover, in public health, there is a distinctly different relationship between provider and recipient, in that in the public health setting, recipients are typically not seeking out the intervention but, rather, are being advised to receive the intervention.
Some have advocated for limited DNA sequence data acquisition and interpretation, modeling the way cystic fibrosis carrier screening is applied, whereas others have advocated for broader data acquisition with limited interpretation. The broader-acquisition approach is founded on the supposition that the net costs of a program in which the number of target conditions expand over time, the way NBS has, will be lower if a single assay is periodically reinterpreted instead of paying for repeat assays. Although the best working model for data acquisition remains to be determined, it seems clear that for population screening the capacity exists to offer interpretation on a very limited number of genes (i.e., Tier 1) at this time.
Such considerations absolutely do not negate the enormous potential for applying genomics in a public health context—but they do imply that a significant measure of caution should be applied as we pursue this exciting prospect. It is clear that at present we do not know how to interpret the results of genome-scale sequencing in the healthy population; our inability to reliably assign pathogenicity to variants, the tendency toward overinterpretation, misinterpretation, and our simple lack of knowledge about the natural history of most genetic disorders argue convincingly that it is premature to implement genome-scale sequencing in healthy individuals outside the realm of research [3, 37-40]. However, we do have a reasonably solid grasp of what the implications are for individuals who carry a pathogenic variation in a few of the approximately 22,000 human genes. This is a good place to begin, with implementation of a targeted sequencing approach that focuses on those few genes that we understand sufficiently to apply to populations.
But even in this context of careful, targeted application, caution is warranted. Lessons learned from population screening should be carefully considered and applied to genomic screening programs. Although there is much to be learned from successes throughout the history of public health, we should also bear in mind examples where applications were prematurely implemented and only later found to cause more harm than good. Thus, as careful programs of targeted sequencing of populations are pursued, we must carefully assess penetrance in the unbiased population setting and collect data on health outcomes to be sure that those screened benefit from the process. We will need comprehensive data on costs to be sure that such interventions are worth pursuing in the broader context of medicine and society. Finally, we must be cognizant of recipients’ acceptance, understanding, and attitudes toward these interventions and work hard to ensure that access to the fruits of such efforts are broadly available and harms are not disproportionately distributed among the population. The future of public health genomics is bright; let us pursue it wisely and responsibly.
Join the conversation!
![]() Tweet this! New from @theNAMedicine: A Proposed Approach for Implementing Genomics-Based Screening Programs for Healthy Adults: https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
Tweet this! New from @theNAMedicine: A Proposed Approach for Implementing Genomics-Based Screening Programs for Healthy Adults: https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
![]() Tweet this! The future of public health genomics is bright; let us pursue it wisely and responsibly. https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
Tweet this! The future of public health genomics is bright; let us pursue it wisely and responsibly. https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
![]() Tweet this! Genomic screening as to improve public health could be an exciting use of the technology. But caution is warranted to ensure that the benefits are broadly available and do not exacerbate disparities: https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
Tweet this! Genomic screening as to improve public health could be an exciting use of the technology. But caution is warranted to ensure that the benefits are broadly available and do not exacerbate disparities: https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
![]() Tweet this! Ethical considerations should loom large when contemplating large-scale genomic screening as a potential public health tool: https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
Tweet this! Ethical considerations should loom large when contemplating large-scale genomic screening as a potential public health tool: https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
![]() Tweet this! Large-scale genomic screening is not only a question of technology – we must also consider the economic, ethical, and societal impacts: https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
Tweet this! Large-scale genomic screening is not only a question of technology – we must also consider the economic, ethical, and societal impacts: https://doi.org/10.31478/201812a #NAMPerspectives #GenomicsRT
Download the graphics below and share them on social media!



References
- Wilson, J. M., Y. G. Jungner, and World Health Organization. 1968. Principles and practice of mass screening for disease. Geneva: World Health Organization. Available at: http://apps.who.int/iris/handle/10665/37650 (accessed November 19, 2018)
- Evans, J. P., J. S. Berg, A. F. Olshan, T. Magnuson, and B. K. Rimer. 2013. We screen newborns, don’t we? Realizing the promise of public health genomics. Genetics in Medicine 15:332-334. https://doi.org/10.1038/gim.2013.11
- Evans, J. P., B. C. Powell, and J. S. Berg. 2017. Finding the rare pathogenic variants in a human genome. JAMA 317(18):1904-1905. https://doi.org/10.1001/jama.2017.0432
- Lindor, N. M., S. N. Thibodeau, and W. Burke. 2017. Whole-genome sequencing in healthy people. Mayo Clinic Proceedings 92(1):159-172. https://doi.org/10.1016/j.mayocp.2016.10.019
- American College of Medical Genetics and Genomics (ACMG). 2018. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Available at: https://www.ncbi.nlm.nih.gov/clinvar/docs/acmg (accessed August 6, 2018).
- Kalia, S. S., K. Adelman, S. J. Bale, W. K. Chung, C. Eng, J. P. Evans, G. E. Herman, S. B. Hufnagel, T. E. Klein, B. R. Korf, K. D. McKelvey, K. E. Ormond, C. S. Richards, C. N. Vlangos, M. Watson, C. L. Martin, and D. T. Miller. 2017. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genetics in Medicine 19(2):249-255. https://doi.org/10.1038/gim.2016.190
- Centers for Disease Control and Prevention (CDC). 2014. Tier 1 genomics applications and their importance to public health. Available at: https://www.cdc.gov/genomics/implementation/toolkit/tier1.htm (accessed June 15, 2018).
- Richards, S., N. Aziz, S. Bale, D. Bick, S. Das, J. Gastier-Foster, W. W. Grody, M. Hegde, E. Lyon, E. Spector, K. Voelkerding, and H. L. Rehm. 2015. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine 17(5):405-424. https://doi.org/10.1038/gim.2015.30
- National Heart, Lung, and Blood Institute (NHLBI). 2011. Expert panel on integrated guidelines for cardiovascular health and risk reduction in children and adolescents: Summary report. Available at: https://www.nhlbi.nih.gov/files/docs/peds_guidelines_sum.pdf (accessed October 23, 2017).
- King, M. C., E. Levy-Lahad, and A. Lahad. 2014. Population-based screening for BRCA1 and BRCA2: 2014 Lasker Award. JAMA 312(11):1091-1092. https://doi.org/10.1001/jama.2014.12483
- Newborn Screening Task Force. 2000. Serving the family from birth to the medical home. Pediatrics 106:383-427. Available at: https://pubmed.ncbi.nlm.nih.gov/10947680/ (accessed September 2, 2020).
- National Institute of Child Health and Human Development (NICHD). 2017. How many newborns are screened in the United States? Available at: https://www.nichd.nih.gov/health/topics/newborn/conditioninfo/infants-screened (accessed September 20, 2018).
- Khoury, M. J., M. S. Bowen, W. Burke, R. J. Coates, N. F. Dowling, J. P. Evans, and J. St. Pierre. 2011. Current priorities for public health practice in addressing the role of human genetics in improving population health. American Journal of Preventive Medicine 40(4):486-493. https://doi.org/10.1016/j.amepre.2010.12.009
- CDC. 2017. The public health system and the 10 essential public health services. Available at: https://www.cdc.gov/stltpublichealth/publichealthservices/essentialhealthservices.html (accessed October 23, 2017).
- Enthoven, A. C. 2009. Integrated delivery systems: The cure for fragmentation. American Journal of Managed Care 15(10 Suppl):S284-290. Available at: https://pubmed.ncbi.nlm.nih.gov/20088632/ (accessed September 2, 2020).
- Gillies, R. R., K. E. Chenok, S. M. Shortell, G. Pawlson, and J. J. Wimbush. 2006. The impact of health plan delivery system organization on clinical quality and patient satisfaction. Health Services Research 41(4 Pt 1):1181-1191. https://doi.org/10.1111/j.1475-6773.2006.00529.x
- Armitage, G. D., E. Suter, N. D. Oelke, and C. E. Adair. 2009. Health systems integration: State of the evidence. International Journal of Integrated Care 9:e82. Available at: https://www.ijic.org/article/10.5334/ijic.316/ (accessed September 2, 2020).
- Weir, H. K., R. N. Anderson, S. M. Coleman King, A. Soman, T. D. Thompson, Y. Hong, B. Moller, and S. Leadbetter. 2016. Heart disease and cancer deaths─trends and projections in the United States, 1969-2020. Preventing Chronic Disease 13:E157. Available at: https://www.cdc.gov/pcd/issues/2016/16_0211.htm (accessed September 2, 2020).
- Ray, T. 2017. Air Force sequencing study to inform broader genomic medicine implementation among service members. GenomeWeb News. Available at: https://www.genomeweb.com/genetic-research/air-force-sequencing-study-inform-broader-genomic-medicine-implementation-among (accessed October 25, 2017).
- De Castro, M., L. G. Biesecker, C. Turner, R. Brenner, C. Witkop, M. Mehlman, C. Bradburne, and R. C. Green. 2016. Genomic medicine in the military. NPJ Genomic Medicine 1:15008. https://doi.org/10.1038/npjgenmed.2015.8
- Buchanan, A. H., A. K. Rahm, and J. L. Williams. 2016. Alternate service delivery models in cancer genetic counseling: A mini-review. Frontiers in Oncology 6:120. https://doi.org/10.3389/fonc.2016.00120
- Carey, D. J., S. N. Fetterolf, F. D. Davis, W. A. Faucett, H. L. Kirchner, U. Mirshani, M. F. Murray, D. T. Smelser, G. S. Gerhard, and D. H. Ledbetter. 2016. The Geisinger MyCode community health initiative: An electronic health record–linked biobank for precision medicine research. Genetics in Medicine 18(9):906-913. https://doi.org/10.1038/gim.2015.187
- Akard, T. F., S. Wray, and M. J. Gilmer. 2015. Facebook advertisements recruit parents of children with cancer for an online survey of web-based research preferences. Cancer Nursing March-April; 38(2):155-161. https://doi.org/10.1097/NCC.0000000000000146
- Martinez, O., E. Wu, A. Z. Shultz, J. Capote, J. López Rios, T. Sandfort, J. Manusov, H. Ovejero, A. Carballo-Dieguez, S. Chavez Baray, E. Moya, J. López Matos, J. J. DelaCruz, R. H. Remien, and S. D. Rhodes. 2014. Still a hard-to-reach population? Using social media to recruit Latino gay couples for an HIV intervention adaptation study. Journal of Medical Internet Research April 24;16(4):e113. https://doi.org/10.2196/jmir.3311
- Sturm, A. C. 2016. Cardiovascular cascade genetic testing: Exploring the role of direct contact and technology. Frontiers in Cardiovascular Medicine 3:11. https://doi.org/10.3389/fcvm.2016.00011
- Roberts, M. C., W. D. Dotson, C. S. DeVore, E. M. Bednar, D. J. Bowen, T. G. Ganiats, R. Fisk Green, G. M. Hurst, A. R. Philip, C. N. Ricker, A. C. Sturm, A. M. Trepanier, J. L. Williams, H. A. Zierhut, K. A. Wilemon, and H. Hampel. 2018. Delivery of cascade screening for hereditary conditions: A scoping review of the literature. Health Affairs 37(5):801-808. https://doi.org/10.1377/hlthaff.2017.1630
- NICHD. 2017. Who pays for newborn screening? Available at: https://www.nichd.nih.gov/health/topics/newborn/conditioninfo/how-used/pages/pays.aspx (accessed December 18, 2017).
- Amendola, L. M., M. O. Dorschner, P. D. Robertson, et al. 2015. Actionable exomic incidental findings in 6503 participants: Challenges of variant classification. Genome Research 25(3):305-315. https://doi.org/10.1101/gr.183483.114
- Adams, M. C., J. P. Evans, G. E. Henderson, and J. S. Berg. 2016. The promise and peril of genomic screening in the general population. Genetics in Medicine 18(6):593-599. https://doi.org/10.1038/gim.2015.136
- Louter, L., J. Defesche, and J. Roeters van Lennep. 2017. Cascade screening for familial hypercholesterolemia: Practical consequences. Supplements 30:77-85. https://doi.org/10.1016/j.atherosclerosissup.2017.05.019
- Morgenstern, J., R. A. Hegele, and J. Nisker. 2015. Simple genetics language as source of miscommunication between genetics researchers and potential research participants in informed consent documents. Public Understanding of Science 24(6):751-766. https://doi.org/10.1177/0963662514528439
- Robinson, J. O., M. J. Slashinski, T. Wang, S. G. Hilsenbeck, and A. L. McGuire. 2013. Participants’ recall and understanding of genomic research and large-scale data sharing. Journal of Empirical Research on Human Research Ethics 8(4):42-52. https://doi.org/10.1525/jer.2013.8.4.42
- Shahu, A., J. Schwartz, M. Perez, S. M. Bernheim, H. M. Krumholz, and E. S. Spatz. 2017. Discerning quality: An analysis of informed consent documents for common cardiovascular procedures. BMJ Quality and Safety 26(7):569-571. https://doi.org/10.1136/bmjqs-2016-005663
- Simonds, V. W., E. M. Garroutte, and D. Buchwald. 2017. Health literacy and informed consent materials: Designed for documentation, not comprehension of health research. Journal of Health Communication 22(8):682-691. https://doi.org/10.1080/10810730.2017.1341565
- Caulfield, T., and B. Murdoch. 2017. Genes, cells, and biobanks: Yes, there’s still a consent problem. PLoS Biology 15(7):e2002654. https://doi.org/10.1371/journal.pbio.2002654
- Rothstein, M. A., J. T. Wilbanks, and K. B. Brothers. 2015. Citizen science on your smartphone: An ELSI research agenda. Journal of Law, Medicine, and Ethics 43(4):897-903. https://doi.org/10.1111/jlme.12327
- Corpas, M., W. Valdivia-Granda, N. Torres, B. Greshake, A. Coletta, A. Knaus, A. P. Harrison, M. Cariaso, F. Moran, F. Nielsen, D. Swan, D. Y. Weiss Solís, P. Krawitz, F. Schacherer, P. Schols, H. Yang, P. Borry, G. Glusman, P. N. Robinson. 2015. Crowdsourced direct-to-consumer genomic analysis of a family quartet. BMC Genomics 16:910. https://doi.org/10.1186/s12864-015-1973-7
- Haggerty, C. M., C. A. James, H. Calkins, C. Tichnell, J. B. Leader, D. N. Hartzel, C. D. Nevius, S. A. Pendergrass, T. N. Person, M. Schwartz, M. D. Ritchie, D. J. Carey, D. H. Ledbetter, M. S. Williams, F. E. Dewey, A. Lopez, J. Penn, J. D. Overton, J. G. Reid, M. Lebo, H. Mason-Suares, C. Austin-Tse, H. L. Rehm, B. P. Delisle, D. J. Makowski, V. C. Mehra, M. F. Murray, and B. K. Fornwalt. 2017. Electronic health record phenotype in subjects with genetic variants associated with arrhythmogenic right ventricular cardiomyopathy: A study of 30,716 subjects with exome sequencing. Genetics in Medicine 19(11):1245-1252. https://doi.org/10.1038/gim.2017.40
- Van Driest, S. L., Q. S. Wells, S. Stallings, W. S. Bush, et al. 2016. Association of arrhythmia-related genetic variants with phenotypes documented in electronic medical records. JAMA 5;315(1):47-57. https://doi.org/10.1001/jama.2015.17701
- Vassy, J. L., K. D. Christensen, E. F. Schonman, C. L. Blout, J. O. Robinson, J. B. Krier, P. M. Diamond, M. Lebo, K. Machini, D. R. Azzariti, D. Dukhovny, D. W. Bates, C. A. MacRae, M. F. Murray, H. L. Rehm, A. L. McGuire, R. C. Green, and MedSeq Project. 2017. The impact of whole-genome sequencing on the primary care and outcomes of healthy adult patients: A pilot randomized trial. Annals of Internal Medicine June 27;167(3):159-169. https://doi.org/10.7326/M17-0188